
 |
Viewmol is hosted by
|
| |||
| Overview | Getting Viewmol | Documentation | Examples | Screenshots | |
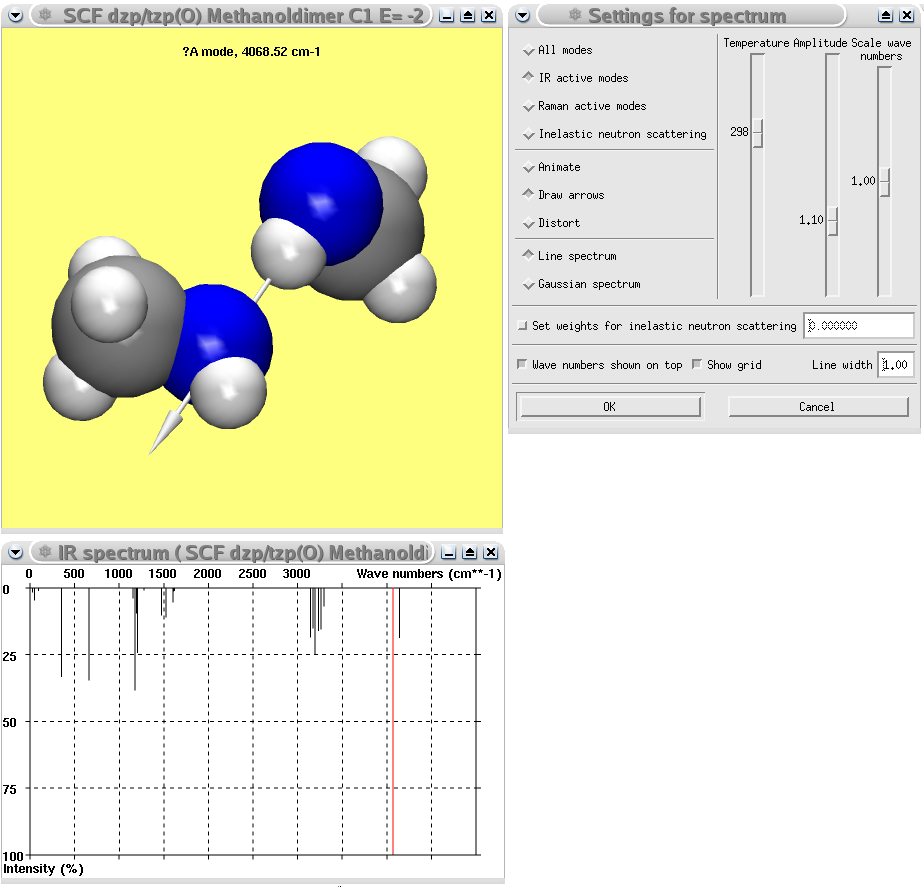
A Gaussian calculation of the methanol dimer showing the calculated IR spectrum and the normal coordinates of the most intense mode.
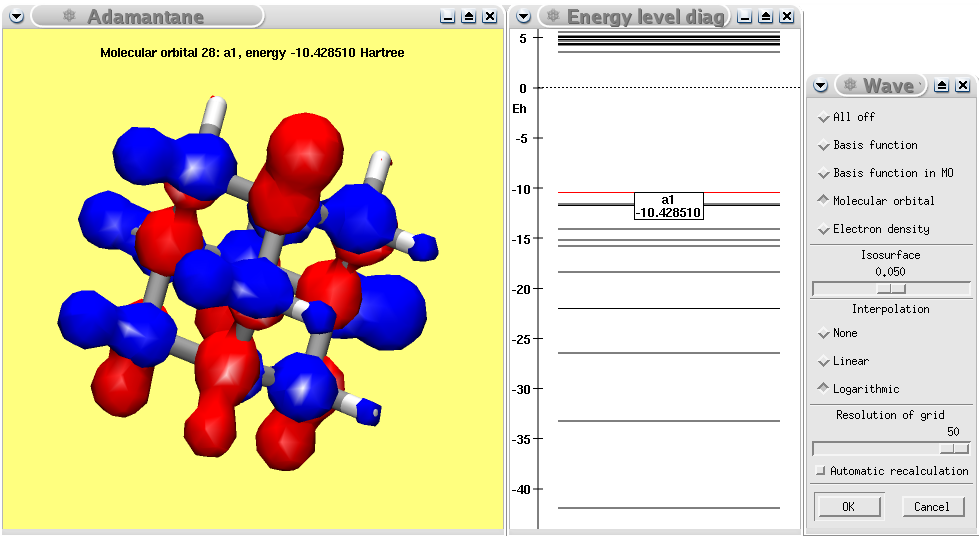
A MOPAC calculation of adamantane showing the calculated HOMO.
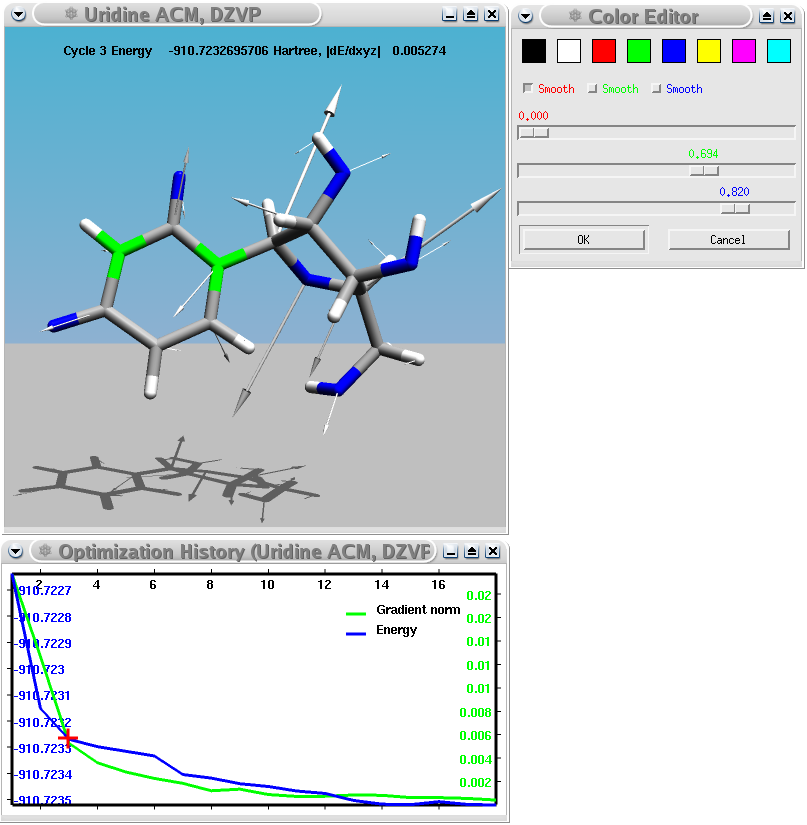
A Turbomole geometry optimization of the uridine nucleoside showing energy and gradient norm as function of the number of iterations and the forces acting on each atom in one iteration.
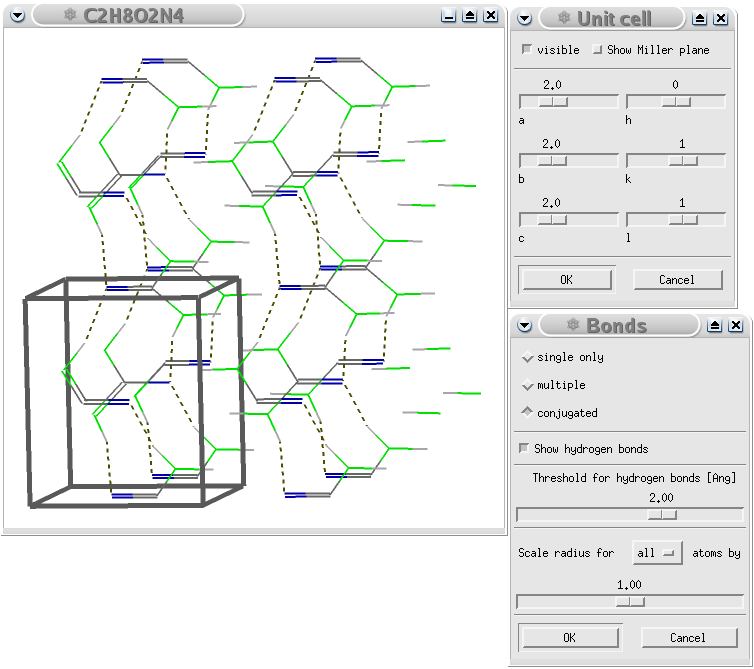
A GULP calculation of urea showing the hydrogen bond pattern in the crystal. Note that the additional cells have been created by Viewmol.
| Jörg-Rüdiger Hill, Tue Mar 9 08:03:10 CET 2004 |